Looking for something?
Can Oligonucleotides Target RNA Splicing to Treat Disease?
The human body is phenomenally complex, and we are continually discovering incredible facts about it. One fascinating aspect lies in splicing of RNA as it is processed in the cell. Alternate splicing of a single pre-mRNA molecule can create tens of thousands of different mRNAs, which contributes to the existence of such an intricate organism as a human. If we could harness the various splicing mechanisms, and catalogue all the possible mRNAs generated, it would have far-reaching outcomes for human health. Thanks to dedicated teams, oligonucleotide therapeutics have already allowed us to correct splicing defects to treat disease.
What is the significance of RNA splicing in human diseases?
First, it is important to understand the profound impact of splicing on the expression of genes and biological pathways.
Inside the cell nucleus, DNA is transcribed into the mRNA template that codes for a protein. The mRNA is then exported to the cytoplasm where it is translated, and the cell makes the protein. However, inside the nucleus complex processes occur to create the mRNA. What is actually transcribed from DNA is called precursor messenger RNA (pre-mRNA). This pre-mRNA is then spliced to create mature mRNA.
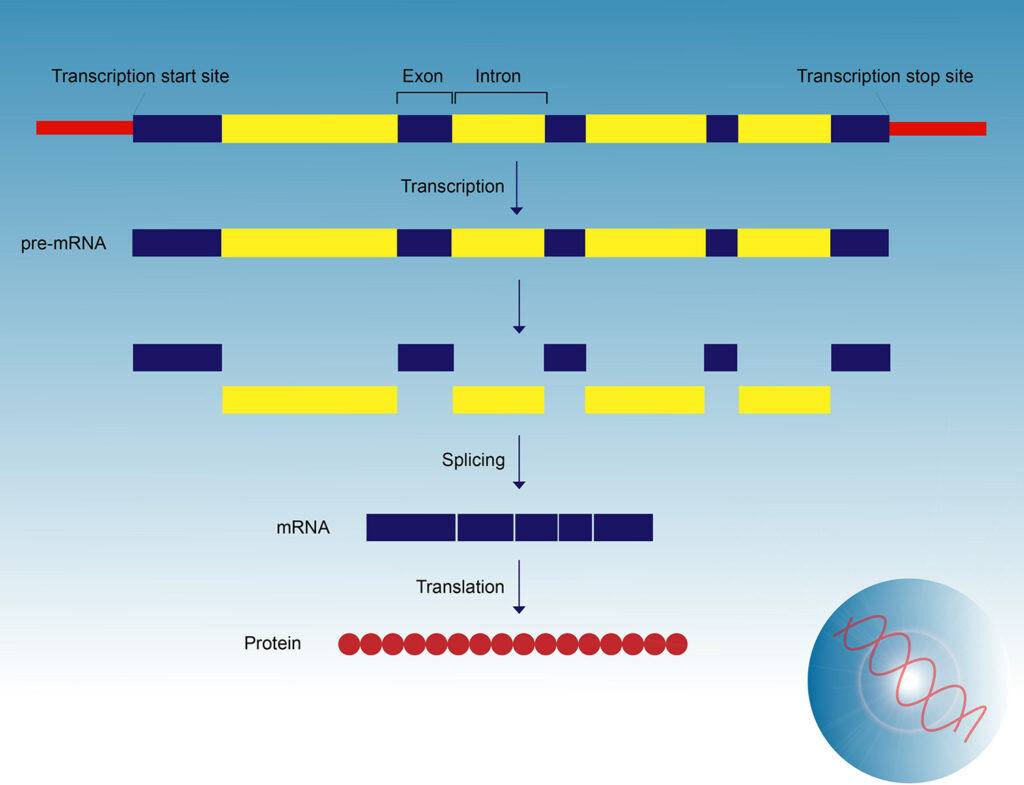 Pre-mRNA must undergo splicing because genes that code protein contain coding sequences (exons) that are interspersed with non-coding sequences (introns). During splicing, introns are removed and exons are joined together before the mRNA is transported out of the nucleus. Without accurate splicing, most protein coding genes cannot be properly expressed.
Pre-mRNA must undergo splicing because genes that code protein contain coding sequences (exons) that are interspersed with non-coding sequences (introns). During splicing, introns are removed and exons are joined together before the mRNA is transported out of the nucleus. Without accurate splicing, most protein coding genes cannot be properly expressed.
The splicing process is highly orchestrated and involves many components and reactions. We will briefly explain just a few.
Splicing is catalyzed by the spliceosome, a complex macromolecular machine made up of hundreds of proteins and a core of five small nuclear RNAs (snRNAs). The job of a spliceosome is to recognize the splice sites, cut out the introns, and splice together the flanking exons (1). The average gene contains 9-10 introns but may have over 100 (2). Each human cell contains around 100,000 spliceosomes, and they remove over 200,000 different intron sequences (2, 3).
However, the splicing process is even more elaborate. One of the most fascinating things about this process is that a single pre-mRNA can be spliced to generate multiple mRNAs, resulting in many different possible proteins (4). This is called alternative splicing (AS) and 95-100% of multiexon genes undergo alternative splicing (5, 6). Alternative splicing usually involves skipping an exon. It may also involve using different 5’ splice sites (ss) or 3’ ss (a 5’ss at one end of an intron and a 3’ss at the other), intron retention, and mutually exclusive exons (4, 7, 8).
The process is further controlled by splicing silencers and enhancers, which are regulatory elements that recruit RNA binding proteins to activate or repress adjacent splice sites (1, 4). They act in conjunction with highly conserved splicing signals (9). Exonic splicing enhancers promote inclusion of the exon they are in, while exonic splicing silencers inhibit its inclusion. When found in an intron, the splicing enhancers and inhibitors do not work upon the intron. Instead, they enhance or inhibit the usage of adjacent exons or splice sites (8).
Splicing must be done with extreme accuracy, as faulty splicing causes the formation of aberrant protein or the loss of normal protein expression (7), resulting in many different diseases and cancers (1, 4).
There are approximately 21,000 protein coding genes in the human genome and many different mutations can cause splicing problems. Non-productive splicing events have been identified in 7,757 protein coding genes, and 1,246 of these are associated with disease (10).
As you can imagine, the potential for error is astronomical, given the sheer number of genes, introns, and exons in each protein coding gene, and the intricate processes involved in creating mature mRNA. It is a wonder that most humans do not have diseases due to splicing errors. However, it would be ideal to eliminate all splicing related genetic diseases and cancers. If therapeutic applications could be developed to target non-productive splicing events, vast improvement could be made in the health and lives of millions of people around the world who suffer from the diseases that result.
ASOs alter splicing to treat disease.
Dominski and Kole first demonstrated that ASOs could be used to target splicing in 1993. Beta-thalassemia is a blood disorder in which low levels of hemoglobin are produced and it is caused by a defect in splicing due to a mutation in the beta-globin gene. They showed that this aberrant splicing could be blocked and redirected to the correct splice site using a splice-switching antisense oligonucleotide (SSO). This was the first time that antisense oligonucleotides were used to restore the function of a defective gene (11).
Splice-switching antisense oligonucleotides (SSOs) are short, modified nucleic acids that target and alter splicing in a therapeutic manner (3). After administration, SSOs enter the cell where they move into the nucleus to affect pre-mRNA splicing (3). SSOs work differently than conventional ASOs, which inhibit gene expression by degrading target pre-mRNA (4).
The first clinical trials testing SSOs in humans evaluated treatment of Duchenne Muscular Dystrophy (DMD). Two SSO approaches were pursued; Eteplirsen by Sarepta Therapeutics, which received FDA approval in 2016, and Kyndrisa pursued by Biomarin which has been withdrawn. The SSOs that have been tested to treat DMD induce skipping of various DMD exons (3).
SSOs are an ideal drug because they are easy to synthesize and deliver, are highly target specific, exhibit widespread entry into most cell types in the body, are well-tolerated even in the CNS, and have a long-lasting effect. They can also be designed to inhibit or enhance a specific splice site, resulting in many different effects on gene expression (3). “ASOs, and in particular SSOs, offer a treatment approach that allows for specific and defined control of gene expression that can be easily tailored to correct or bypass the effects of a specific mutation” (3).
A second approach to altering splicing through the use of ASOs has been highly successful in the treatment of spinal muscular atrophy (SMA). SMA is caused by mutations in the SMN1 gene that codes for the SMN protein. The SMN2 gene acts as a backup but there is a slight difference in the SMN2 sequence. This difference causes an unbalance between splicing silencers and enhancers of exon 7, causing exon 7 to be excluded. Therefore, it only produces low amounts of a shorter SMN protein and cannot completely compensate for the lack of SMN1. However, patients with a higher number of copies of SMN2 have a less severe form of the disease, providing an avenue of treatment (3).
Spinraza (nusinersen) was the first drug approved by the FDA to treat spinal muscular atrophy. It is an antisense oligonucleotide (ASO) that binds to the main splicing silencer of exon 7, modulating alternative splicing of the SMN2 gene, which increases the levels of functional SMN protein in the central nervous system (1). Ionis Pharmaceuticals partnered with Biogen to deliver Spinraza after it was developed by Ionis in collaboration with Drs. Adrian Krainer, Yimin Hua, and colleagues at Cold Spring Harbor Laboratory (CSHL).
Patients with spinal muscular atrophy who are treated with Spinraza display significant improvements in motor skills and muscle function as well as stable disease activity (3). An interim analysis showed just how wildly successful Spinraza is. Most infants with type 2 SMA are never able to walk, and those with type 1 need permanent ventilation and nearly always die before their second birthday. In the trial, children who were most likely to develop SMA type 1 and 2 were treated before symptom onset and before turning 6 weeks old. After 4.8 years, all the children were alive and did not need permanent ventilation! 96 percent of the children were also able to walk with assistance, and many were able to walk independently in the normal timeframe. These phenomenal results display the potential in using oligonucleotides to treat splicing defects.
Stoke Therapeutics has recently tested another interesting approach in mice called targeted augmentation of nuclear gene output (TANGO) that uses ASOs to modulate splicing. It reduces non-productive alternative splicing to increase productive mRNA, which leads to an increase of full-length and fully functional protein, slowing or stopping disease progression. TANGO may offer another gene-specific approach to treat the underlying cause of severe genetic disease (10).
Other therapeutic approaches are in development.
Splicing-based therapeutic approaches fall under two main categories, correcting aberrant splicing and inducing aberrant splicing. Approaches to correct aberrant splicing include correcting: splice site mutations, cryptic and pseudo splice sites, mutations that disrupt splicing regulatory sequences, and dysregulated alternative splicing. Aberrant splicing can be induced to knock down detrimental gene expression and rescue gene expression through RNA reframing (7). In addition to ASOs, various technologies are in development to harness the approaches.
Bifunctional oligonucleotides are able to redirect splicing. They are made of an antisense portion that determines target specificity and a non-hybridizing tail that recruits proteins or RNA/protein complexes that affect splice site selection through silencing (TOSS, targeted oligonucleotide silencer of splicing) or enhancing (TOES, targeted oligonucleotide enhancer of splicing) (4, 12). Bifunctional TOSS/TOES restores protein expression by redirecting splicing resulting in exon inclusion (4).
Small interfering RNAs (siRNAs) can be utilized to induce degradation of aberrant and alternatively spliced mRNAs by targeting exonic/intronic sequences. This method is especially useful in targeting cancers, as specific splice variants are increased during tumorigenesis. Not only does utilizing siRNAs downregulate certain tumor associated proteins, but it also increases apoptosis, decreases cell proliferation, and increases the sensitivity of tumor cells to chemotherapeutic agents (4).
Spliceosome-mediated RNA trans-splicing (SMaRT) rescues full-length gene expression by replacing the entire coding sequence upstream or downstream of a target splice site to repair RNA, producing a full-length, functional RNA (4, 7). It utilizes a pre-trans-splicing molecule (PTM) that consists of an ASO to target the intron of the mutated splice site, a synthetic splice site, and an RNA sequence that is spliced to the endogenous RNA (7). This technology has been tested in models of spinal muscular atrophy, cystic fibrosis, hemophilia A, retinitis pigmentosa, tauopathies, and frontotemporal dementia with parkinsonism-17, and results show that it can successfully reprogram gene expression (4).
Spliceosomal small nuclear RNAs (snRNAs) are a component of the spliceosome that are essential for the splicing of introns. Modified snRNAs could potentially be used to eliminate the skipping of some exons and correct mutation induced splice defects (4). Modified snRNAs have been tested in models of Haemophilia B, cystic fibrosis, and spinal muscular atrophy (SMA) (13), and a range of other disease-causing mutations (4).
Many other therapeutic approaches are also being developed to correct splice-related defects. Amongst these are small molecule compounds and genome editing approaches that include zinc finger proteins (ZFPs), transcription activator-like effector nucleases (TALENs), and CRISPR-Cas9 (4).
Databases, Artificial Intelligence, and innovative Technologies are providing rapid discovery of splicing errors and the means to develop new therapies.
The previously mentioned developments by Stoke Therapeutics, Sarepta Therapeutics, Ionis Pharmaceuticals, and Biogen represents just a small portion of the work being conducted in this field. Ionis and Cold Spring Harbor Laboratory continue to work together. Recently, they have developed multiple effective methods to significantly reduce off-target effects of splice-switching ASOs (14).
Sarepta built upon their pioneering work with phosphorodiamidate morpholino oligomer (PMO) chemistries that were the basis of directing splicing to skip exons and provided therapies for Duchenne muscular dystrophy and currently have many other PMOs in their development pipeline. Peptide phosphorodiamidate morpholino oligomers (PPMOs) are Sarepta’s next-generation chemistry that adds a peptide to the PMO structure, with the goal of increasing cell nuclei penetration.
Amylon has developed RNA Modulation Technology which uses ASOs to alter the splicing of pre-mRNA to counteract the effects of disease-causing mutations. They are using their RNA Modulation Technology to target rare genetic disorders of the central nervous system (CNS) and develop life-changing therapeutic solutions for patients with serious unmet medical needs.
The Hastings Lab at Chicago Medical School – Rosalind Franklin University studies how pre-mRNA splicing and expression of small non-coding regulatory microRNAs are altered in diseased cells. They are developing approaches to treat Batten disease using antisense oligonucleotides to manipulate pre-mRNA splicing. CLN3 Batten disease is often caused by a deletion encompassing exons 7 and 8, resulting in a reading frame-shift. Recently, Centa, et al. used an ASO to induce skipping of exon 5 of CLN3 to restore the open reading frame. They discovered that “a single treatment of neonatal mice with an exon 5-targeted ASO-induced robust exon skipping for more than a year, improved motor coordination, reduced histopathology in Cln3Δex7/8 mice and increased survival in a new mouse model of the disease. ASOs also induced exon skipping in cell lines derived from patients with CLN3 Batten disease (15).”
An ASO has previously been successfully used to treat Batten Disease in the case of Mila Makovec, a child who was diagnosed with a rare, fatal form of Batten disease. Milasen, an ASO targeting splicing of a gene called MFSD8, was the first personalized ASO approved by the FDA. Milasen was developed by Dr. Timothy Yu and a large team of colleagues.
Dr. Timothy Yu is the principal investigator of The Yu Lab at Boston Children’s Hospital. The primary mission of Dr. Yu and other members of the lab is to investigate neurodevelopmental and neurogenetic diseases through translational genomics. They also use translational genomic medicine to study and develop antisense therapies for rare neurological diseases, many of which are caused by aberrant splicing. Another interesting area of research by Yu’s team is the study of ‘human gene knockouts’ in autism.
Envisagenics uses artificial intelligence and a database of around seven million RNA splicing errors to link errors to diseases, providing rapid discovery of splicing errors. This allows them to discover new drug targets and design therapeutics to correct them.
Biogen and Envisagenics recently announced a collaboration to advance ribonucleic acid (RNA) splicing research within central nervous system (CNS) diseases. Through the collaboration, Biogen believes they will be able to use Envisagenics’ SpliceCore platform and database of splicing errors to identify and validate genetic targets of disease to increase the probability of success in CNS drug discovery.
Envisagenics has also entered into a research program agreement with Johnson & Johnson’s Lung Cancer Initiative (LCI). They will analyze LCI’s data using their SpliceCore platform and build predictive models for lung cancer progression and risk in the hopes of identifying RNA splicing events that can predict lung cancer, and eventually develop new therapies for patients.
An intriguing prospect.
Pre-RNA splicing is a fascinating and beautifully intricate process. However, when something goes wrong it can result in deadly diseases. Discovering which defects lead to specific disease states and harnessing this process may prove to be the key to treating many genetic diseases and cancers.
References:
- Abramowicz A, Gos M. (2018). Splicing mutations in human genetic disorders: examples, detection, and confirmation. Journal of applied genetics, 59(3), 253–268. https://doi.org/10.1007/s13353-018-0444-7
- Chen W, Moore MJ. Spliceosomes. Curr Biol. 2015 Mar 2;25(5):R181-3. doi: 10.1016/j.cub.2014.11.059. PMID: 25734262.
- Havens, M. A., & Hastings, M. L. (2016). Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic acids research, 44(14), 6549–6563. https://doi.org/10.1093/nar/gkw533
- Suñé-Pou, M., Limeres, M. J., Moreno-Castro, C., Hernández-Munain, C., Suñé-Negre, J. M., Cuestas, M. L., & Suñé, C. (2020). Innovative Therapeutic and Delivery Approaches Using Nanotechnology to Correct Splicing Defects Underlying Disease. Frontiers in genetics, 11, 731. https://doi.org/10.3389/fgene.2020.00731
- Pan Q., Shai O., Lee L. J., Frey B. J., Blencowe B. J. (2008). Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Genet.40 1413–1415. 10.1038/ng.259
- Wang E. T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., et al. (2008). Alternative isoform regulation in human tissue transcriptomes. Nature456 470–476. 10.1038/nature07509
- Havens, M. A., Duelli, D. M., & Hastings, M. L. (2013). Targeting RNA splicing for disease therapy. Wiley interdisciplinary reviews. RNA, 4(3), 247–266. https://doi.org/10.1002/wrna.1158
- Wang, Z., & Burge, C. B. (2008). Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA (New York, N.Y.), 14(5), 802–813. https://doi.org/10.1261/rna.876308
- Mount, S.M. (2005). Exonic splicing enhancers and exonic splicing silencers. In Encyclopedia of Genetics, Genomics, Proteomics and Bioinformatics (eds L.B. Jorde, P.F.R. Little, M.J. Dunn and S. Subramaniam). https://doi.org/10.1002/047001153X.g402314
- Lim, K. H., Han, Z., Jeon, H. Y., Kach, J., Jing, E., Weyn-Vanhentenryck, S., Downs, M., Corrionero, A., Oh, R., Scharner, J., Venkatesh, A., Ji, S., Liau, G., Ticho, B., Nash, H., & Aznarez, I. (2020). Antisense oligonucleotide modulation of non-productive alternative splicing upregulates gene expression. Nature communications, 11(1), 3501. https://doi.org/10.1038/s41467-020-17093-9
- Dominski, Z., & Kole, R. (1993). Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proceedings of the National Academy of Sciences of the United States of America, 90(18), 8673–8677. https://doi.org/10.1073/pnas.90.18.8673
- Brosseau, J. P., Lucier, J. F., Lamarche, A. A., Shkreta, L., Gendron, D., Lapointe, E., Thibault, P., Paquet, E., Perreault, J. P., Abou Elela, S., & Chabot, B. (2014). Redirecting splicing with bifunctional oligonucleotides. Nucleic acids research, 42(6), e40. https://doi.org/10.1093/nar/gkt1287
- Fernandez Alanis, E., Pinotti, M., Dal Mas, A., Balestra, D., Cavallari, N., Rogalska, M. E., Bernardi, F., & Pagani, F. (2012). An exon-specific U1 small nuclear RNA (snRNA) strategy to correct splicing defects. Human molecular genetics, 21(11), 2389–2398. https://doi.org/10.1093/hmg/dds045
- Scharner, J., Ma, W. K., Zhang, Q., Lin, K. T., Rigo, F., Bennett, C. F., & Krainer, A. R. (2020). Hybridization-mediated off-target effects of splice-switching antisense oligonucleotides. Nucleic acids research, 48(2), 802–816. https://doi.org/10.1093/nar/gkz1132
- Centa, J. L., Jodelka, F. M., Hinrich, A. J., Johnson, T. B., Ochaba, J., Jackson, M., Duelli, D. M., Weimer, J. M., Rigo, F., & Hastings, M. L. (2020). Therapeutic efficacy of antisense oligonucleotides in mouse models of CLN3 Batten disease. Nature medicine, 26(9), 1444–1451. https://doi.org/10.1038/s41591-020-0986-1