Presenter: Keith Gagnon, Southern Illinois University, USA
Date: March 21, 2019
Description:
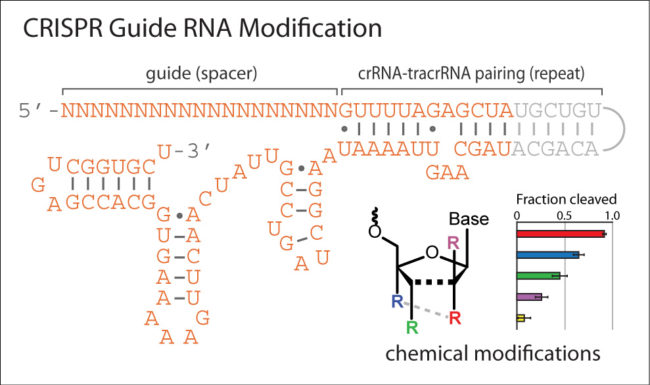
CRISPR-based genome editing has emerged as an exciting technology for biomedical and therapeutic applications. Since 2015, a number of studies have explored the effects of chemically modifying the guide RNA components of CRISPR enzymes and the spectrum of modifications that are compatible. While primarily focused on improving editing efficiency or reducing off-target effects, these investigations have led to some important rules regarding the placement and type of modifications that may be used. This webinar will discuss the progress in this area of research and provide practical guidelines for chemically modifying CRISPR RNA.
Recording of the Webinar: Click Play to View
In addition, Dr. Gagnon would like to make the slides available for download: Gagnon OTS webinar
Presenter Biography

Keith Gagnon, Southern Illinois University, USA
Dr. Keith T. Gagnon is an Assistant Professor at Southern Illinois University School of Medicine, USA since 2014 and was elected to the OTS Board of Directors in 2018.
He obtained a BS in Biochemistry (2003) and a PhD in RNA biochemistry (2007) from North Carolina State University. From 2008-2014 he worked as a Post-Doc with Dr. David Corey, where he characterized mechanisms of nuclear RNAi, its necessary cellular machinery and potential for therapeutic application. He also investigated siRNAs and ASOs as therapeutic modalities for repeat expansion disorders, including Huntington’s disease, spinocerebellar ataxia 3, and C9ORF72-associated frontotemporal dementia and amyotrophic lateral sclerosis.
His laboratory focuses on two areas: neurological repeat expansion disorders and RNA-guided enzymes. For the latter, his research focuses on the design, development, and control of CRISPR-based proteins for therapeutic applications, including chemical modification and RNA-protein engineering.






