
January 2020 Joint Papers of the Month
Directed RNase H Cleavage of Nascent Transcripts Causes Transcription Termination
Lai F, Damle SS, Ling KK, Rigo F.
Mol Cell. 2020 Jan 3. pii: S1097-2765(19)30959-1.
Joint First Authors
Antisense-Mediated Transcript Knockdown Triggers Premature Transcription Termination.
Lee JS, Mendell JT.
Mol Cell. 2020 Jan 2. pii: S1097-2765(19)30924-4.

Prof. Joshua Mendell and Dr. Jong-Sun Lee (l to r)
A unified allosteric/torpedo mechanism for transcriptional termination on human protein-coding genes.
Eaton JD, Francis L, Davidson L, West S.
Genes Dev. 2020 Jan 1;34(1-2):132-145.

Joshua D. Eaton and Prof. Steven West
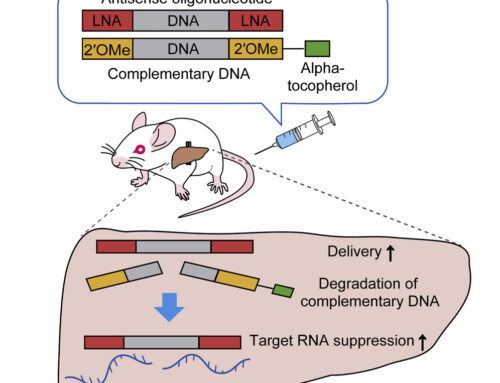
RNaseH-mediated antisense oligonucleotides (ASOs), or gapmers, are widely used in therapeutic and research applications. One particular research application is determining if a long noncoding RNAs (lncRNAs) has an RNA- or transcription-dependent function.
For a refresher on the molecular cell biology of gapmers watch Prof. Stanley Crooke’s Lifetime Achievement Award lecture:
Lai and colleagues from Ionis Pharmaceuticals designed a series of 5-10-5 MOE-PS ASOs targeted to intron or exon sequences of six different genes. Treatment with all ASOs resulted in dose-dependent reductions in target mRNA levels. Equally strong reductions in pre-mRNA levels were only seen with intron targeted ASOs and when measured using PCR primers proximal to or downstream of the target site. Chromatin immunoprecipitation (ChIP) showed reduced Pol II occupancy downstream of the ASO target site with intron-targeted gapmers showing more pronounced reductions. ASOs targeted to the last exon were functional without inducing transcription termination. This was dependent on exonuclease XRN2 and thus due to the torpedo mechanism of transcription termination.
Lee and Mendell show similar results for lncRNAs. Using nuclear run-off assays combined with qRT-PCR, they also demonstrate that nascent transcript levels are significantly reduced. ChIP confirmed that this was due to reductions in Pol II occupancy at the 3′ end of transcripts and siRNA-mediated XRN2 depletion returned these levels to normal.
Importantly, only ASOs targeting sequences at the 3′ end of transcripts mediate RNA degradation without concomitant reduction in transcription.
Eaton and colleagues focus on the mechanistic details of transcription termination, but also show that ASOs can induce premature termination and discuss the implications of that for lncRNA research.
Why you should read it
Careful design of ASOs targeting lncRNAs is necessary to distinguish between their RNA- and transcription-dependent functions. Re-evaluation of the published lncRNA literature with these findings in mind is essential.