
Base-pair Conformational Switch Modulates miR-34a Targeting of Sirt1 mRNA
Baronti L, Guzzetti I, Ebrahimi P, Friebe Sandoz S, Steiner E, Schlagnitweit J, Fromm B, Silva L, Fontana C, Chen AA, Petzold K.

Lorenzo Baronti
As part of RNA-induced silencing complexes (RISCs), microRNAs bind to target mRNAs and regulate their expression post-transcriptionally. Initial binding between guide and target RNA is mediated by the microRNA seed sequence, with at least 5 nucleotides starting from the 2nd position (g2-g7) base pairing with perfect complementarity to the target mRNA.
For more detail on microRNAs and the seed sequence, please see this lecture by Prof. Bartel:
Published data suggests that the nature of the downstream binding between the two RNAs, including bulges and mismatches, determines if the target mRNA is cleaved or not. However, the detailed mechanism is unclear. To study this, Baronti and colleagues from the Karolinska Institute and University at Albany use R1ρ relaxation dispersion nuclear magnetic resonance combined with molecular simulations.
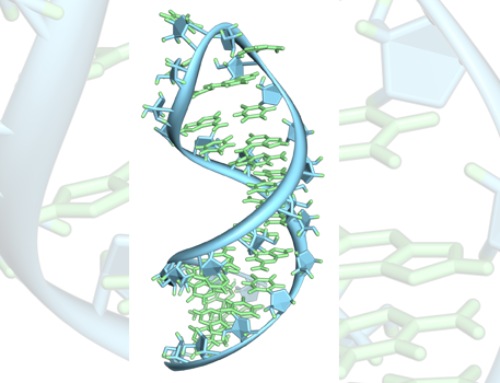
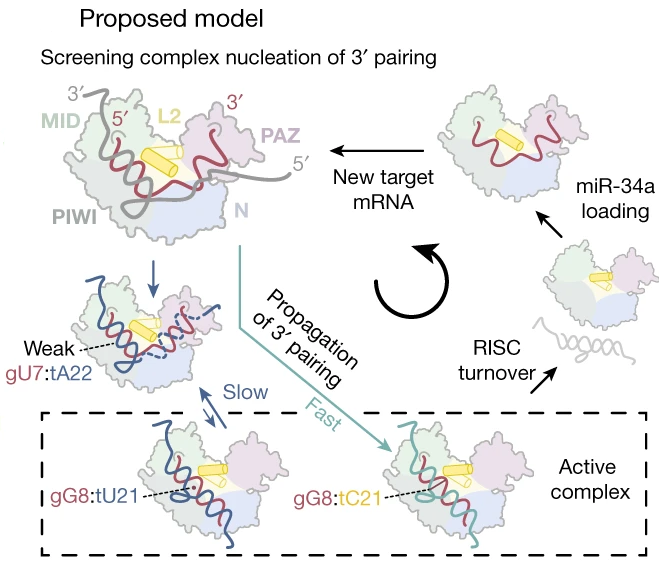
When studying the binding of human miR-34a-5p to its target Sirt1, the authors found that there is an equilibrium between the predominant but weak 7-mer-A1 seed (position g1-g7) and a secondary 8-mer seed sequence. The extended seed forms when the G at position 8 switches from binding C17 in the Sirt1 mRNA to forming a G:U wobble base pair with U21. This forces the bulge in the Sirt1 mRNA to bend at 87° instead of 146°. A mutated miR-34a trapped into forming the extended 8-mer seed sequence adopted a similarly angled conformation with no effect on melting temperature, dissociation constant or binding affinity of the duplex. However, the mutated miR-34a increased target downregulation by two-fold.
To investigate how the changed RNA duplex confirmation fits within the Ago2 binding site, the authors used slow-growth simulations based on existing crystal structures. These simulations show that the changed angle reorients the microRNA 3′end from within the PAZ-domain to along the PIWI and N domains of Ago2. This leads to opening of the RISC associated with the active form. Five other targets of miR-34a selected based on a sequence search and secondary-structure prediction also showed a 50–80% increase in target-downregulation when forced to form the 8-mer seed.
Based on this data, the authors “suggest a model whereby RISC undergoes a structural transition mediated by RNA dynamics: the RISC first screens targets for correct seed pairing, than transitions into an active complex, releasing the miR-34a 3′-end which is allowed to fully bind the Sirt1 mRNA in the compensatory region.”
Why you should read it
Studying the conformational dynamics of microRNA-target duplexes instead of simple sequence alignments should result in improved guide RNA design.